Tutorial: Using MATLAB PCT and Parallel Server in Red Cloud
Introduction
This tutorial introduces you to various ways in which you can use the MATLAB Parallel Computing Toolbox (PCT) to offload your computationally intensive work from your MATLAB client to Red Cloud. You do this through the MATLAB Parallel Server (formerly called the MATLAB Distributed Computing Server or MDCS, in releases prior to R2019a).
In order to run the examples in this tutorial, you must have completed the steps in MATLAB Parallel Server in Red Cloud. Use those instructions to launch an appropriate Red Cloud instance of at least 2 cores which will serve as your MATLAB Parallel Server "cluster". The Parallel Server cluster should be able to pass the built-in validation test in your local MATLAB client, as explained on that page.
Overview of PCT and Parallel Server
What is MATLAB Parallel Server?
Quoting from The MathWorks:
- MATLAB Parallel Server™ lets you scale MATLAB® programs and Simulink® simulations to clusters and clouds.
- Prototype and debug applications on the desktop with Parallel Computing Toolbox™, and easily scale to clusters or clouds without recoding.
- Parallel Server supports batch jobs, interactive parallel computations, and distributed computations with large matrices. You can use the MATLAB optimized scheduler provided with MATLAB Parallel Server.
Parallel resources: local and remote
Use the Parallel menu to choose the cluster where your PCT parallel workers ("labs") will run. Often this will be the local cluster, which is merely the collection of processor cores on the machine where your MATLAB client is running. (Local is the default unless you say otherwise.) However, you can also specify a remote cluster that has been made available to you through MATLAB Parallel Server.
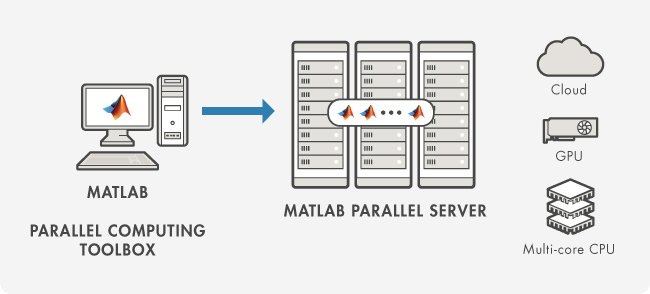
PCT opens up parallel possibilities
MATLAB has multithreading built into its core libraries, but multithreading mostly aids big array operations and is not within user control. The Parallel Computing Toolbox (PCT) enables user-directed parallelism that allows you to take better advantage of the available CPUs.
-
PCT supports various kinds of parallel computations that you can run interactively...
- Parallel for-loops: parfor
- Single program, multiple data: spmd, pmode
- Array partitioning for big-data parallelism: (co)distributed
-
PCT also facilitates batch-style parallelism...
- Multiple independent runs of a serial function: createJob
- Single run of parallelized code: createCommunicatingJob
This tutorial focuses mainly on batch-style usage, because such computations do not tie up the console of your MATLAB client. It is therefore a suitable way to offload work to Red Cloud--which is most often seen as a distinct resource for handling long-running jobs. But along the way, you will also get an overview of how parallel programming works generally in PCT. For more in-depth coverage, you can refer to The MathWorks' own documentation.
Two ways to use PCT, interactive vs. batch-style
- Interactive: Start local or remote pool using parpool, run PCT commands directly (can be scripted)
- Batch: Specify local or remote cluster using parcluster, submit jobs and task functions to the cluster
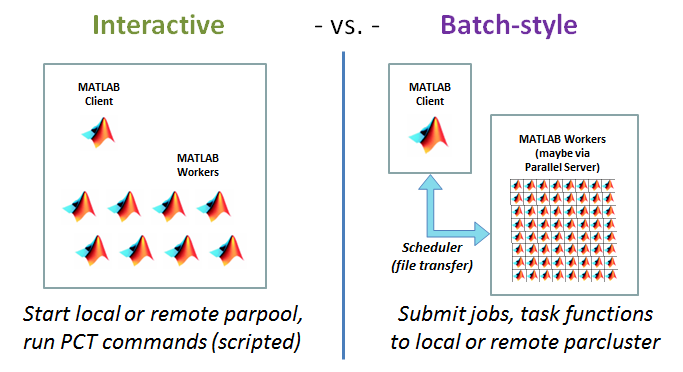
Interactive PCT: Major Concepts
The MATLAB documentation presents a wide array of options for parallelizing your code with PCT. The parallel execution strategy depends on the specific commands that are chosen. We'll start with commands that would be more typical of interactive work. The interactive pool of workers can exist on your local laptop, or on a Parallel Server cluster like Red Cloud.
- parpool: pool of distinct MATLAB processes = "labs"
- Labs are aware of each other; they can work together
- Differs from multithreading! No shared address space
- This is ultimately what allows the same basic concepts to work on remote, distributed-memory clusters
- parfor: parallel for-loop, iterations must be independent
- Labs (workers) in the pool split up iterations
- Communication among labs is needed because load balancing is built in
- spmd: single program, multiple data
- All labs in the pool execute every command
- Programmer specifies how and when the labs communicate
- (co)distributed: array is partitioned among labs
- distributed from client's point of view; codistributed from labs' point of view
- Treated as "multiple data" for spmd, but appears as one array to MATLAB functions
The MathWorks provides a complete list of PCT functions and details on how they are used.
Built-in functions that can use a parpool automatically
Certain MATLAB functions will use an existing parpool if one is available. Examples may be found in the Global Optimization toolbox (e.g., fmincon), the Statistics and Machine Learning toolbox, and others. To make it happen, simply set the 'UseParallel' option to 'true' or 'always' or 1. Check the MathWorks documentation for details. This is an easy method for enabling parallel processing when your computations are concentrated into just one or two built-in functions.
Batch-Style PCT: Basic Workflow
Within a MATLAB session, submitting jobs to a MATLAB Parallel Server in Red Cloud follows a general pattern:
-
The user designates a Red Cloud instance to be the parallel "cluster" (parcluster).
- This allows jobs to be submitted to the MATLAB Job Scheduler (MJS) on that instance for execution, rather than the local machine.
-
Individual jobs are created, configured, and submitted by the user.
- Jobs may be composed of one or more individual tasks.
- If tasks within the job consist of one's own MATLAB code, the function files will be auto-uploaded to the cluster by default.
- Data files may be included in the upload to the cluster by specifying them in the AttachedFiles cell array.
- Alternatively, files can be uploaded in advance to Red Cloud using typical clients such as sftp or scp. MATLAB Parallel Server finds them through the AdditionalPaths cell array.
-
Job state and results are saved in Red Cloud and synced back to the client.
- Job state and results are retained until deleted by the user using the delete(job) function, or until the instance is terminated.
- Job state and results from past and present jobs can be viewed from any client at any time.
- The exact ending time of a job depends on the nature of the tasks that have been defined within it and the available resources.
- PCT has callbacks that allow actions to be taken upon job completion, depending on completion status.
Jobs and tasks
- parcluster creates a cluster object, which allows you to create Jobs.
- In PCT, Jobs are containers for Tasks, which are where the actual work is defined.
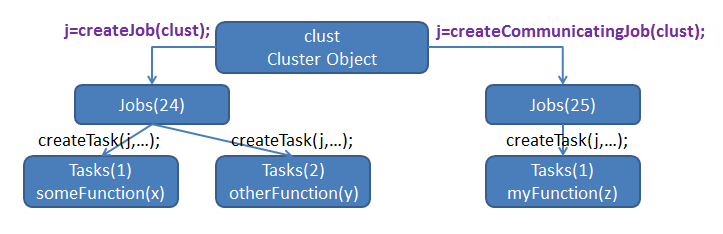
Types of jobs
Batch-style PCT has 3 types of jobs: independent, SPMD, and pool.
-
Independent: createJob()
- Can contain many tasks -- possibly more than the number of available workers
- Workers continue to run tasks in parallel, one per worker, until all tasks are done
-
SPMD: createCommunicatingJob(...,'Type','SPMD',...)
- Has ONE task to be run by ALL workers, just like a spmd block
- Typically makes use of explicit message passing syntax such as labBarrier, labindex, etc.
- Could also define one or more codistributed arrays for data too large to fit into the memory of any one machine
-
Pool: createCommunicatingJob(...,'Type','Pool',...)
- Has ONE task which is run by ONE worker
- Other workers run spmd blocks or parfor loops in the task
- Mimics the interactive mode of using PCT
Experience has shown that independent jobs work the best for running parametric sweeps. SPMD jobs could serve for this, but execution would be more synchronized than necessary. Perhaps a pool job seems most natural for doing this, especially if you are accustomed to running parametric sweeps using a parfor loop and a local pool. But in a pool job, one worker is essentially wasted, as it does nothing more than manage the loop iterations of the other workers. Therefore, for Red Cloud, consider converting your code from the parfor style to a collection of independent jobs and tasks; it should be a relatively easy process.
This tutorial will cover only very simple examples of independent, SPMD, and pool job submission. If you need help with any of the commands please be sure to make use of MATLAB's built-in help function.
Simple independent job
For this example we’ll consider a trivial function that takes any input and waits 5 seconds before returning the input value. We will run this function locally, then compare with distributed execution in Red Cloud.
First, create a file simple_independent_job.m containing the following:
function output = simple_independent_job(input)
pause(5);
output = input;
end
Running simple_independent_job in your local MATLAB window should yield (after a pause):
>> simple_independent_job(1)
ans =
1
To run it on MATLAB Parallel Server, we will use the createJob function. Here is an example of running the same code on a 2-core instance in Red Cloud. We assume that the instance is already validated, and that it is the default cluster in the Parallel menu.
>> clust = parcluster();
>> job = createJob(clust);
>> task = createTask(job, @simple_independent_job, 1, {1});
>> submit(job);
>> wait(job);
>> fetchOutputs(job)
ans =
cell
[1]
This by itself is not that interesting, but let's pretend we want to run simple_independent_job 10x in a row. Running locally in serial, this should take 10x5 = 50 seconds.
>> tic; for i=1:10; simple_distributed_job(1); end; toc
Elapsed time is 50.012614 seconds.
But if we run it as a PCT independent job, we can make it finish faster. Note that we don't have to upload a copy of the "simple_independent_job.m" file to the Red Cloud instance in advance because the AutoAttachFiles property is true (logical value 1) by default.
>> job = createJob(clust);
>> for i=1:10
createTask(job,@simple_independent_job, 1, {i});
end
>> tic; submit(job); wait(job); toc
Elapsed time is 34.117942 seconds.
The independent job takes 30% less time to run than it did in serial. Since there are 2 workers, one might expect a speedup of 2 rather than 1.43, but there is an overhead associated with time to submit the job and retrieve the results from the remote server. Calling fetchOutputs on the job object yields the results of the 10 tasks.
>> fetchOutputs(job)
ans =
10×1 cell array
[ 1]
[ 2]
[ 3]
[ 4]
[ 5]
[ 6]
[ 7]
[ 8]
[ 9]
[10]
Simple SPMD job
A SPMD job is meant to be run in parallel on multiple workers that know about each other and can therefore work as a team. The workers ("labs") can even communicate with each other using an MPI-like syntax, summarized below.
For this example, we won't go into the details of inter-worker communication. We will merely demonstrate that each worker receives a unique index that can be used to assign unique work to it. Here's a trivial function that illustrates the role of labindex:
function output = simple_spmd_job()
output = labindex;
end
Running this function locally just produces the answer "1", because no parallel workers are requested or defined in the function itself. However, if the same function is submitted in a SPMD job, then each worker runs the body of the function and returns a different answer, just as if the body of the function were running inside a MATLAB spmd block. To show this, we'll again assume that a 2-core instance is already running in Red Cloud, and that it is set up to be the default cluster in the Parallel menu:
>> clust = parcluster();
>> job = createJob(clust,'Type','SPMD');
>> task = createTask(job, @simple_spmd_job, 1, {});
>> submit(job);
>> wait(job);
>> fetchOutputs(job)
ans =
2×1 cell array
{[1]}
{[2]}
The above cell array can be converted to a simple array with syntax like a = [ans{:}].
Simple pool job
For this example we'll consider yet another trivial function that pauses 1 second 50 times in a row before returning the input value.
function output = simple_pool_job(input)
for i=1:50
%parfor i=1:50
pause(1);
end
output=input;
end
Running the code locally we might we would expect the function to take 50 seconds to run. What happens if we change the for for-loop into a parfor-loop (by moving the comment sign to the previous line) and run it as a pool job in Red Cloud? Let's assume that our Red Cloud instance with two MATLAB labs is still active and the default profile still points to it:
>> clust = parcluster();
>> job = createCommunicatingJob(clust,'Type','Pool');
>> task = createTask(job, @simple_pool_job, 1, {1});
>> tic; submit(job); wait(job); toc
Elapsed time is 63.563045 seconds.
But wait, running our parfor-loop in a pool job with two labs appears to be far slower than running our original for-loop sequentially! This is because in this example, only 1 worker executes the parfor-loop. The other worker in the cluster acts as a "manager" whose role is to run anything in the main script that isn't in a parfor or spmd section. The parfor-loop thus takes longer, due to the overhead involved in submitting/retrieving the job.
Accordingly, pool jobs only make sense if there are quite a few workers besides the manager to run the parallel tasks.
** Note:** If we were to submit this same job to the local cluster, assuming a 2-core personal workstation, we would get 2 labs for the pool job. The reason for this difference is that the MATLAB GUI is already running on the local machine, so there is no need to assign one worker as a manager. (Try it—but don’t forget to switch back to Red Cloud when you’re done!)
Comparison with using a pool in Red Cloud interactively
Submitting batch jobs to a MATLAB cluster in Red Cloud has the advantage that it frees up the MATLAB client on your workstation to do other tasks. However, there are times when it makes sense to run a fast parallel computation in real time, so that you can get results in (say) minutes instead of hours.
You can use the parpool command to start a pool on your cluster in Red Cloud, after which any PCT commands will be sent to the workers in Red Cloud. In fact, if Red Cloud is your default cluster, then any parfor or spmd block that you enter in your MATLAB client will automatically open a pool using the Red Cloud workers.
The above functions for the SPMD and pool jobs can be used to illustrate how this works. For the SPMD example, replace the function line with spmd, and run the body of the function line-by-line in the client. The function in the pool example can just be run directly as follows:
>> tic; simple_pool_job(1); toc
Starting parallel pool (parpool) using the 'Red Cloud' profile ...
connected to 2 workers.
Analyzing and transferring files to the workers ...done.
Elapsed time is 36.040343 seconds.
>> delete(gcp('nocreate'))
Parallel pool using the 'Red Cloud' profile is shutting down.
Simply issuing the parfor command within the function triggers MATLAB to start a pool of workers on the default cluster, and it then uses the pool to execute the loop. When running in interactive mode, the role of manager is performed by the front-end MATLAB client. Therefore, in this case, there really are two workers to execute the parfor-loop, and it runs almost twice as fast. However, as before, parallel overhead prevents the code from running exactly twice as fast on two labs. The final command closes the pool after running the example.
Simple debugging
Here's an example of a common problem and how to diagnose it. It is very easy to forget to upload one or more files that may be needed by your task. The AutoAttachFiles property can help with this, but it only looks for program files (.m, .p, .mex). If you neglect to list your data file in the AttachedFiles cell array, the result will be something like this:
>> clust = parcluster();
>> job = createJob(clust);
>> createTask(job, @type, 0, {'nonexistent_file.txt'});
>> submit(job);
>> wait(job);
>> job
job =
Job
Properties:
ID: 25
Type: independent
Username: slantz
State: finished
SubmitDateTime: 07-Jun-2017 21:10:59
StartDateTime: 07-Jun-2017 21:10:59
Running Duration: 0 days 0h 0m 2s
NumThreads: 1
AutoAttachFiles: true
Auto Attached Files: List files
AttachedFiles: {}
AdditionalPaths: {}
Associated Tasks:
Number Pending: 0
Number Running: 0
Number Finished: 1
Task ID of Errors: [1]
Task ID of Warnings: []
Note the next-to-last line "Task ID of Errors". This indicates that one of the tasks had an error. In the console, the "1" serves a link to error information. But we can also view details of the error by inspecting the task object associated with the job:
>> job.Tasks(1)
ans =
Task with properties:
ID: 1
State: finished
Function: @type
Parent: Job 25
StartDateTime: 07-Jun-2017 21:10:59
Running Duration: 0 days 0h 0m 2s
Error: File 'nonexistent_file.txt' not found.
Warnings:
You can view just the error message by entering job.Tasks(1).Error.
Another good way to get information for debugging purposes is to examine the console output of one or more of the tasks. To do this, however, you will first need to set the CaptureDiary property of the chosen task(s), prior to job submission:
createTask(job, @type, 0, {'nonexistent_file.txt'},'CaptureDiary', true);
If you also insert a few calls to disp() into your task function before running your job, you might get a better understanding why a task might be failing. Once the first task finishes, you can view its console output by entering job.Tasks(1).Diary.
Long running jobs
Does your MATLAB computation require hours or even days to run? You'll be glad to know you can exit MATLAB completely and re-open it later to check on the status of a job that takes a long time to complete. For the sake of example, let's create a job that waits for 5 minutes, then returns. For this example we will just call the built-in pause function and will not attach or upload any code.
>> clust = parcluster();
>> job = createJob(clust);
>> createTask(job, @pause, 0, {300});
>> submit(job);
>> pause(10); job
job =
Job
Properties:
ID: 26
Type: independent
Username: slantz
State: running
SubmitDateTime: 07-Jun-2017 21:29:49
StartDateTime: 07-Jun-2017 21:29:49
Running Duration: 0 days 0h 0m 19s
NumThreads: 1
AutoAttachFiles: true
Auto Attached Files: List files
AttachedFiles: {}
AdditionalPaths: {}
Associated Tasks:
Number Pending: 0
Number Running: 1
Number Finished: 0
Task ID of Errors: []
Task ID of Warnings: []
Note that the job is in the state "running". Assuming we did not do wait(job), it is now possible to use the MATLAB client for other things, or even to exit MATLAB entirely and start a new MATLAB session later. To retrieve information about the previously submitted job, we use the parcluster and findJob functions to retrieve the job that was running. (Note, the cluster must be up and active in Red Cloud for this to work.) Hopefully we remembered to record the "Job ID" from the previous session...
>> clust = parCluster();
>> job = findJob(clust,'ID',26)
job =
Job
Properties:
ID: 26
Type: independent
Username: slantz
State: running
SubmitDateTime: 07-Jun-2017 21:29:49
StartDateTime: 07-Jun-2017 21:29:49
Running Duration: 0 days 0h 3m 33s
NumThreads: 1
AutoAttachFiles: true
Auto Attached Files: List files
AttachedFiles: {}
AdditionalPaths: {}
Associated Tasks:
Number Pending: 0
Number Running: 1
Number Finished: 0
Task ID of Errors: []
Task ID of Warnings: []
We see that the "Running Duration" has advanced by 3 minutes. If you decide later that you do not want the job to complete you can cancel the job using the cancel function.
>> cancel(job);
>> job
job =
Job
Properties:
ID: 26
Type: independent
Username: slantz
State: finished
SubmitDateTime: 07-Jun-2017 21:29:49
StartDateTime: 07-Jun-2017 21:29:49
Running Duration: 0 days 0h 4m 7s
NumThreads: 1
AutoAttachFiles: true
Auto Attached Files: List files
AttachedFiles: {}
AdditionalPaths: {}
Associated Tasks:
Number Pending: 0
Number Running: 0
Number Finished: 1
Task ID of Errors: [1]
Task ID of Warnings: []
The error in this case is simply a notification that the job was canceled.
>> job.Tasks(1).Error
ans =
ParallelException with properties:
identifier: 'distcomp:task:Cancelled'
message: 'The job was cancelled by user "slantz" on machine "192.168.1.147".'
cause: {}
remotecause: {}
stack: [0×1 struct]
If you forgot the ID you can inspect the Jobs field of the MJS Cluster object that was returned by parcluster.
>> clust.Jobs
ans =
11x1 Job array:
ID Type State FinishDateTime Username Tasks
-----------------------------------------------------------------------------
1 7 independent finished 22-May-2017 15:06:02 slantz 1
2 16 independent finished 02-Jun-2017 15:19:35 slantz 1
3 17 independent finished 04-Jun-2017 21:57:32 slantz 1
4 18 independent finished 04-Jun-2017 21:58:49 slantz 1
5 19 independent finished 04-Jun-2017 21:59:22 slantz 1
6 20 independent finished 04-Jun-2017 22:03:52 slantz 10
7 21 independent finished 04-Jun-2017 22:05:50 slantz 10
8 23 pool finished 05-Jun-2017 21:45:40 slantz 2
9 24 independent finished 07-Jun-2017 21:08:20 slantz 1
10 25 independent finished 07-Jun-2017 21:11:01 slantz 1
11 26 independent finished 07-Jun-2017 21:33:56 slantz 1
More on SPMD jobs and SPMD blocks
- The task function for a SPMD job is like the code contained in a spmd block in a MATLAB script.
- Within the task function, you are responsible for implementing parallelism using "labindex" logic.
- The lab* functions allow workers (labs) to communicate; they act just like MPI message-passing methods.
- labSend(data,dest,[tag]); % point-to-point
- labReceive(source,tag); % datatype, size are implicit
- labReceive(); % take any source
- labBroadcast(source); labBarrier; gop(f,x); % collectives
- (Co)distributed arrays are sliced across workers so huge matrices can be operated on. Collect slices with gather.
Distributing work with parfeval and batch
- createJob() isn't the only way to run independent tasks...
- parfeval() requests the given function be excuted on one worker in a parpool, asynchronously
- batch() does the same on one worker NOT in a parpool
- It creates a one-task job and submits it to a parcluster
- It can also be a one-line method for initiating a pool job
- It works with either a function or a script
- Either can easily be called in a loop over a list of tasks
- Use fetchNext() to collect results as they become available
How to distribute work without PCT or Parallel Server
- Create a MATLAB .m file that takes one or more input parameters (such as the name of an input file)
- Apply the MATLAB C/C++ compiler (mcc), which converts the script to C, then to a standalone executable
- Run N copies of the executable on an N-core Red Cloud instance (or on your own laptop or cluster)
- Make sure each executable receives a different input parameter
- On an interactive Linux machine, run each task in the background (&)
- On a cluster, remember that mpirun can launch non-MPI processes, too
- Matlab runtimes (free!) must be available on all nodes
- For process control, write a master script in Python, say
When Is File Transfer Needed?
- If your workers do not share a disk with your client, and they will require datafiles or other custom files
- Recall the earlier example from the "Simple debugging" section:
>> clust = parcluster();
>> job = createJob(clust);
>> createTask(job, @type, 1, {'nonexistent_file.txt'});
>> submit(job);
- The type function is no problem at all, it's built in
- But "nonexistent_file.txt" does not exist on the remote computer
- We’ll want to transfer this file and get it added to the path
MATLAB can copy files... or you can
- Setting the AutoAttachFiles property tells MATLAB to copy files containing your function definitions
- Use the AttachedFiles cell array to copy any data files or directories the task will need; directory structures are preserved
- Not very efficient, though: file transfer occurs separately for each worker running a task for that particular job
- OK for small projects with a couple of files
- A better-scaling alternative is to copy your files to disk(s) on the remote server(s) in advance
- Use AdditionalPaths to make the files available at run time
GPGPU in MATLAB PCT: Fast and Easy
- Many functions are overloaded to call CUDA code automatically if objects are declared with gpuArray type
- Benchmarking with large 1D and 2D FFTs shows excellent acceleration on NVIDIA GPUs
- MATLAB code changes are trivial
- Move data to GPU by declaring a gpuArray
- Call method in the usual way
g = gpuArray(r); f = fft2(g);
Are GPUs really that simple?
- No. Your application must meet four important criteria.
- Nearly all required operations must be implemented natively for type GPUArray.
- The computation must be arranged so the data seldom have to leave the GPU.
- The overall working dataset must be large enough to exploit 100s of thread processors.
- On the other hand, the overall working dataset must be small enough that it does not exceed GPU memory.
Are GPUs available in Red Cloud?
- Yes! But at the present time, Red Cloud does not have an image that supports both MATLAB Parallel Server and GPUs. (Let us know if you want one!)
- You can try out GPU functionality on your laptop or workstation if it has an NVIDIA graphics card.
- Your graphics card must have a sufficiently high level of compute capability and the latest drivers.
- If it looks like you can scale up, please let CAC know! Red Cloud has NVIDIA Tesla V100s; each of them is capable of 14 teraflop/s.
PCT and Parallel Server: The Bottom Line
- PCT can greatly speed up large-scale computations and the analysis of large datasets
- GPU functionality is a nice addition to the arsenal
- Parallel Server allows parallel workers to run on cluster and cloud resources beyond one’s laptop, e.g., Red Cloud
- Yes, a learning curve must be climbed…
- General knowledge of how to restructure code so that parallelism is exposed
- Specific knowledge of PCT functions
- But speed often matters!